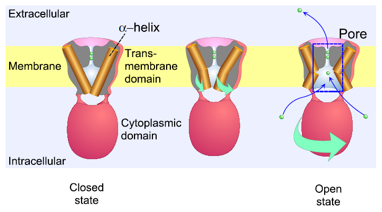
Crystal structure of potassium channels in closed and open states has been elucidated. To gain dynamic pictures of functional channel, the KcsA potassium channel upon gating were examined by the diffracted X-ray tracking (DXT) method. A single molecular KcsA channel was attached with a gold nano-crystal with a size of 20 nm and high flux X-rays were irradiated. A diffraction spot from the nano-crystal was tracked in real time to trace the trajectories of conformational changes. At physiological pH where the channel keeps its gate closed, the channel exhibited small random fluctuations of its structure. When the channel is actively gating at acidic pH, the channel twisted its conformation around the pore axis. Random clockwise and counterclockwise twisting was observed in the range of several tens of degrees. This motion corresponds to the twisting and untwisting of the pore, which allow ions to occlude or permeate. The twisting conformational change was initiated from the transmembrane domain and was propagated towards the end of the cytoplasmic domain. In the presence of an open channel blocker, tetrabutylammonium, the twisting motion was stopped even at acidic pH and the conformational wave failed to propagate. This type of twisting motion may be shared by various types of ion channels upon gating. (H. Shimizu*, M. Iwamoto*, T. Konno*, A. Nihei***, Y. C. Sasaki** & S. Oiki*: Global Twisting Motion of Single Molecular KcsA Potassium Channel upon Gating. Cell 132, 67-78, 2008)
Twisting conformational change of the channel upon gating
The transmembrane domain of the potassium channel is formed by a bundle of α-helices (left). The intracellular ends of the helices are crossed, which occludes the ion permeation pathway. During gating the channel molecule (un)twisted its shape, which corresponds to relaxation of the bundle, leading to opening of the permeation pathway. This twisting motion was originated from the transmembrane domain and was transferred to the cytoplasmic domain.
*Department of Molecular Physiology and Biophysics, University of Fukui Faculty of Medical Sciences, Fukui 910-1193
**SPring-8, JASRI, Hyogo 679-5198
***SII NanoTechnology Inc., Chiba 270-2222